Massive Protein Import into the Early-Evolutionary-Stage Photosynthetic Organelle of the Amoeba Paulinella chromatophora

Mining the Secretome of C2C12 Muscle Cells: Data Dependent Experimental Approach To Analyze Protein Secretion Using Label-Free Quantification and Peptide Based Analysis | Journal of Proteome Research

IJMS | Free Full-Text | Small Molecule Arranged Thermal Proximity Coaggregation (smarTPCA)—A Novel Approach to Characterize Protein–Protein Interactions in Living Cells by Similar Isothermal Dose–Responses

IJMS | Free Full-Text | Small Molecule Arranged Thermal Proximity Coaggregation (smarTPCA)—A Novel Approach to Characterize Protein–Protein Interactions in Living Cells by Similar Isothermal Dose–Responses

Proteome‐Wide Survey of Cysteine Oxidation by Using a Norbornene Probe - Alcock - 2020 - ChemBioChem - Wiley Online Library

IJMS | Free Full-Text | Small Molecule Arranged Thermal Proximity Coaggregation (smarTPCA)—A Novel Approach to Characterize Protein–Protein Interactions in Living Cells by Similar Isothermal Dose–Responses

Solid-phase synthesis of cereblon-recruiting selective histone deacetylase 6 degraders (HDAC6 PROTACs) with anti-leukemic activity | Biological and Medicinal Chemistry | ChemRxiv | Cambridge Open Engage

Proteome-Wide Survey Reveals Norbornene Is Complementary to Dimedone and Related Nucleophilic Probes for Cysteine Sulfenic Acid | Biological and Medicinal Chemistry | ChemRxiv | Cambridge Open Engage

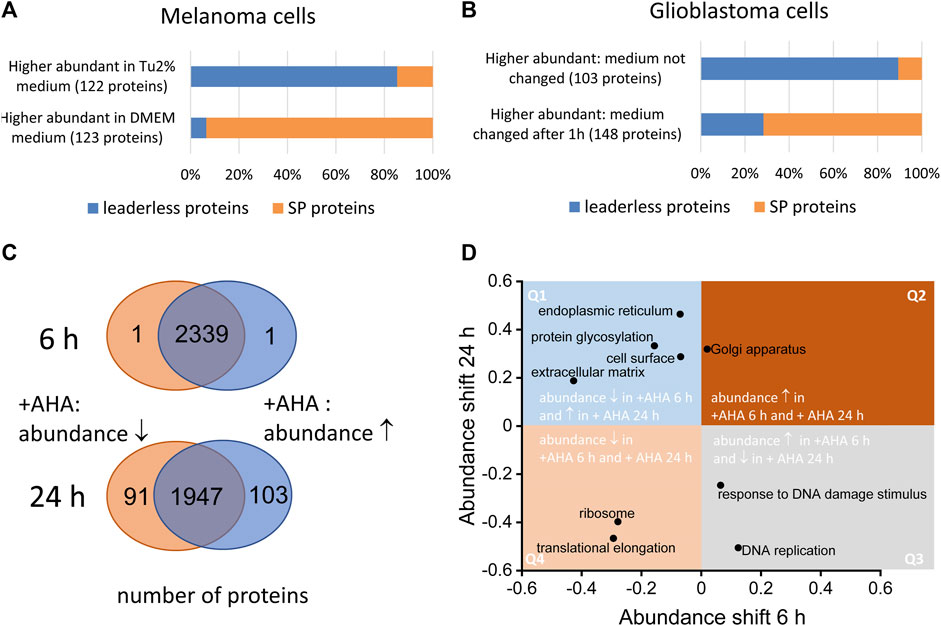
Comparative Secretomics Gives Access to High Confident Secretome Data: Evaluation of Different Methods for the Determination of Bona Fide Secreted Proteins - Poschmann - 2021 - PROTEOMICS - Wiley Online Library
Massive Protein Import into the Early-Evolutionary-Stage Photosynthetic Organelle of the Amoeba Paulinella chromatophora
New Insight into Stimulus-Induced Plasticity of the Olfactory Epithelium in Mus musculus by Quantitative Proteomics

Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics | Biomarkers: A Proteomic Challenge | ScienceDirect.com by Elsevier